Input data and parameters
QualiMap command line
| qualimap bamqc -bam /mnt/galaxy/files/002/252/dataset_2252592.dat -c -nw 400 -hm 3 -sd |
Alignment
| Command line: | bwa mem -t 2 -v 1 localref.fa /mnt/pulsar/files/staging/1348713/inputs/dataset_2252007.dat /mnt/pulsar/files/staging/1348713/inputs/dataset_2252008.dat |
| Draw chromosome limits: | yes |
| Analyze overlapping paired-end reads: | yes |
| Program: | bwa (0.7.17-r1188) |
| Analysis date: | Thu May 14 11:20:45 AEST 2020 |
| Size of a homopolymer: | 3 |
| Skip duplicate alignments: | yes (only flagged) |
| Number of windows: | 400 |
| BAM file: | /mnt/galaxy/files/002/252/dataset_2252592.dat |
Summary
Warnings
| No flagged duplicates are detected | Make sure duplicate alignments are flagged in the BAM file or apply a different skip duplicates mode. |
Globals
| Reference size | 1,193,446 |
| Number of reads | 103,860 |
| Mapped reads | 103,860 / 100% |
| Unmapped reads | 0 / 0% |
| Mapped paired reads | 103,860 / 100% |
| Mapped reads, first in pair | 51,930 / 50% |
| Mapped reads, second in pair | 51,930 / 50% |
| Mapped reads, both in pair | 103,860 / 100% |
| Mapped reads, singletons | 0 / 0% |
| Secondary alignments | 0 |
| Supplementary alignments | 14 / 0.01% |
| Read min/max/mean length | 30 / 125 / 125 |
| Overlapping read pairs | 3,181 / 6.13% |
| Duplicated reads (flagged) | 0 / 0% |
| Duplicated reads (estimated) | 4,680 / 4.51% |
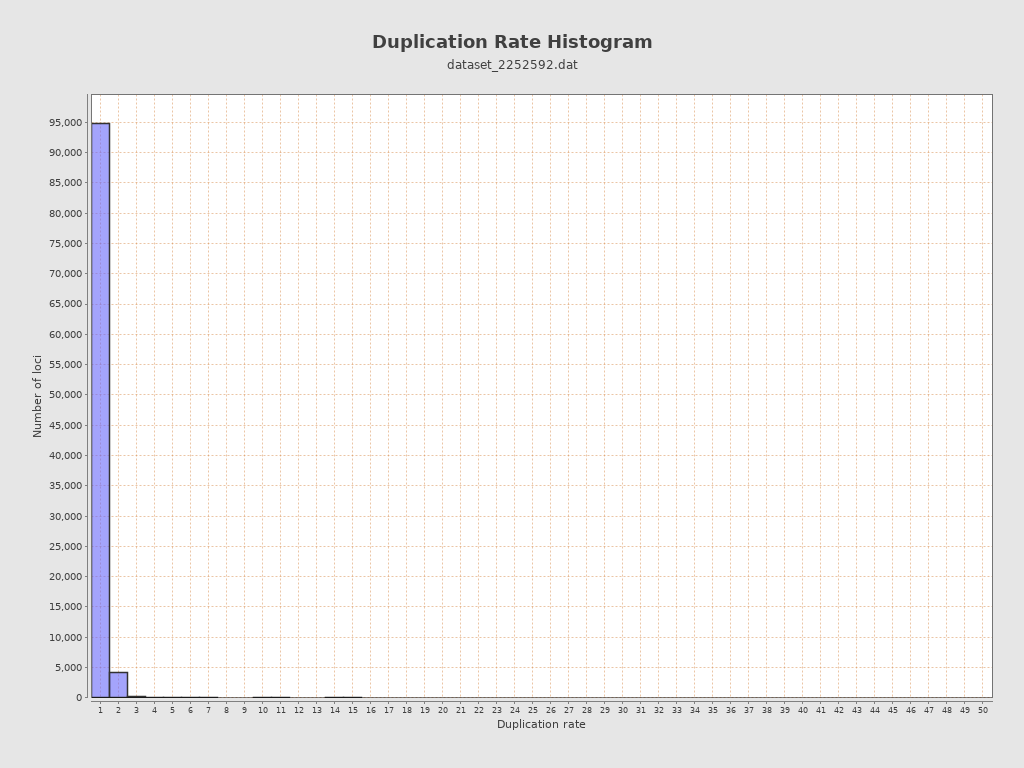
| Duplication rate | 4.46% |
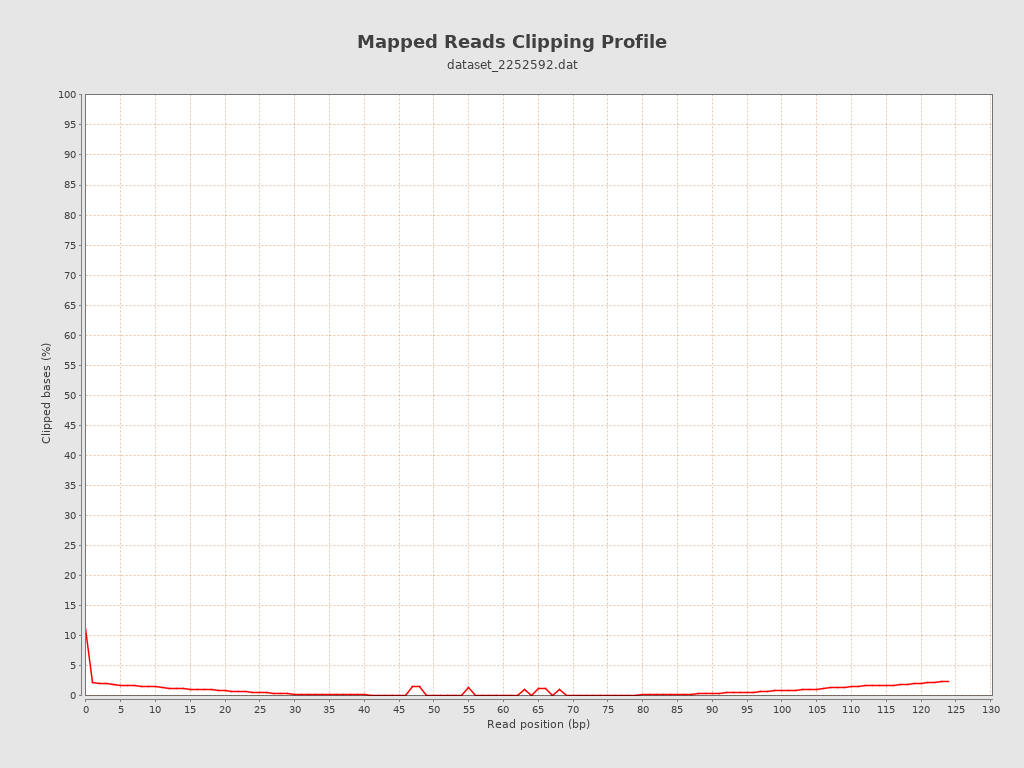
| Clipped reads | 245 / 0.24% |
ACGT Content
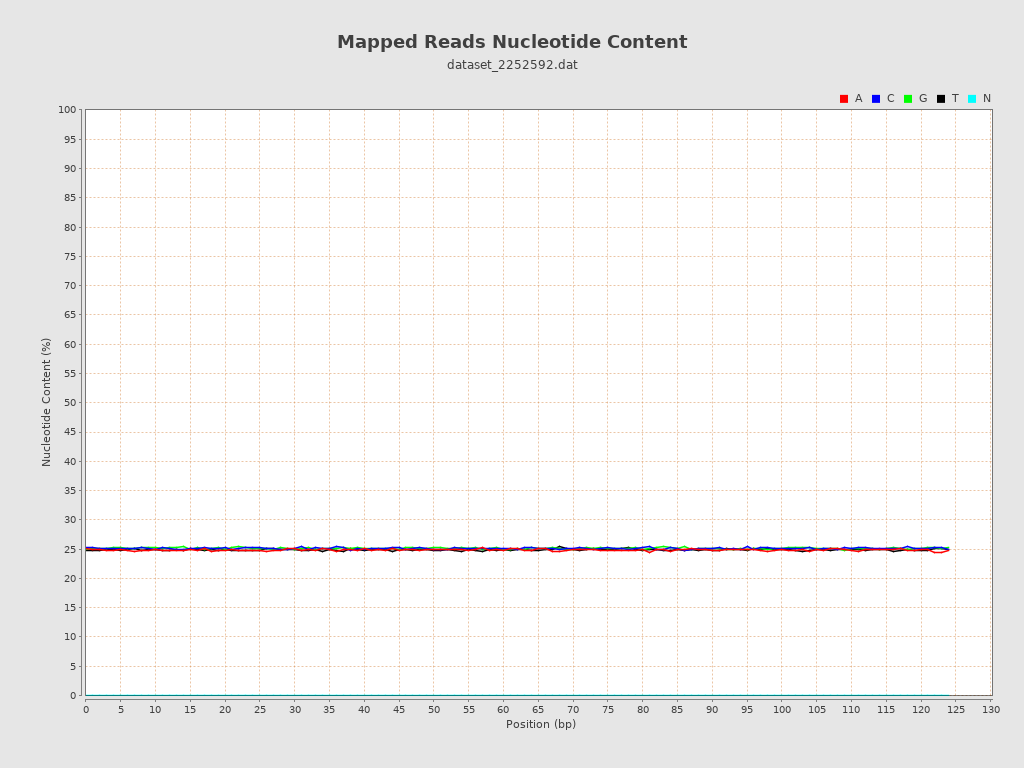
| Number/percentage of A's | 3,226,338 / 24.86% |
| Number/percentage of C's | 3,260,286 / 25.12% |
| Number/percentage of T's | 3,229,525 / 24.88% |
| Number/percentage of G's | 3,262,360 / 25.14% |
| Number/percentage of N's | 0 / 0% |
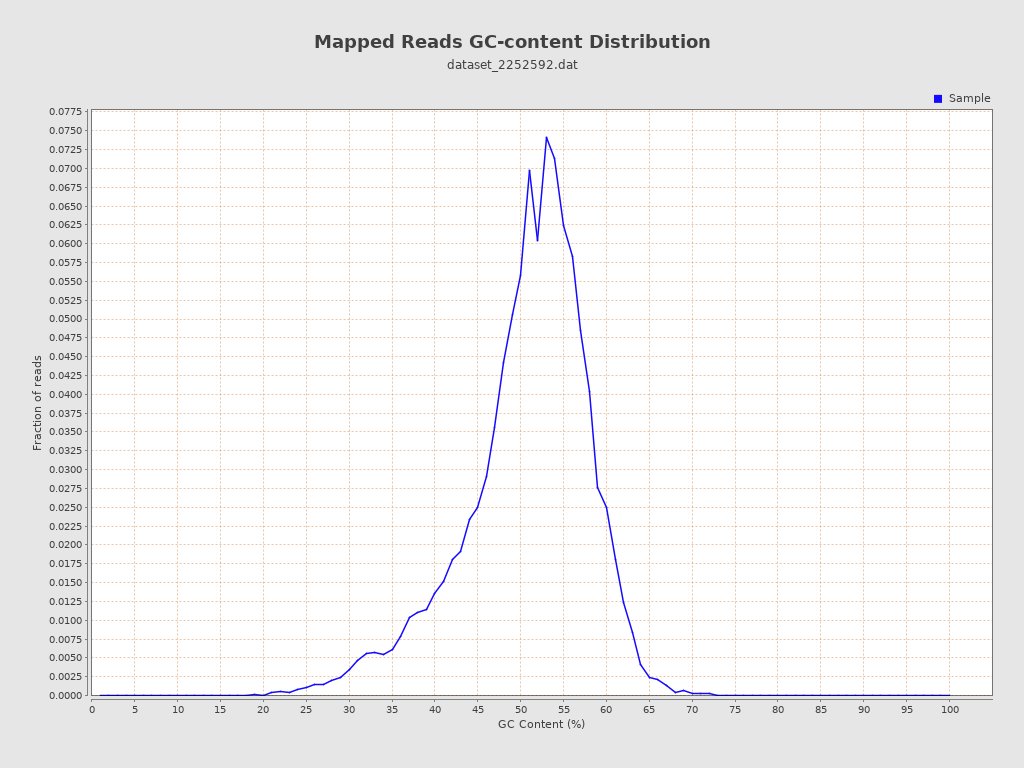
| GC Percentage | 50.26% |
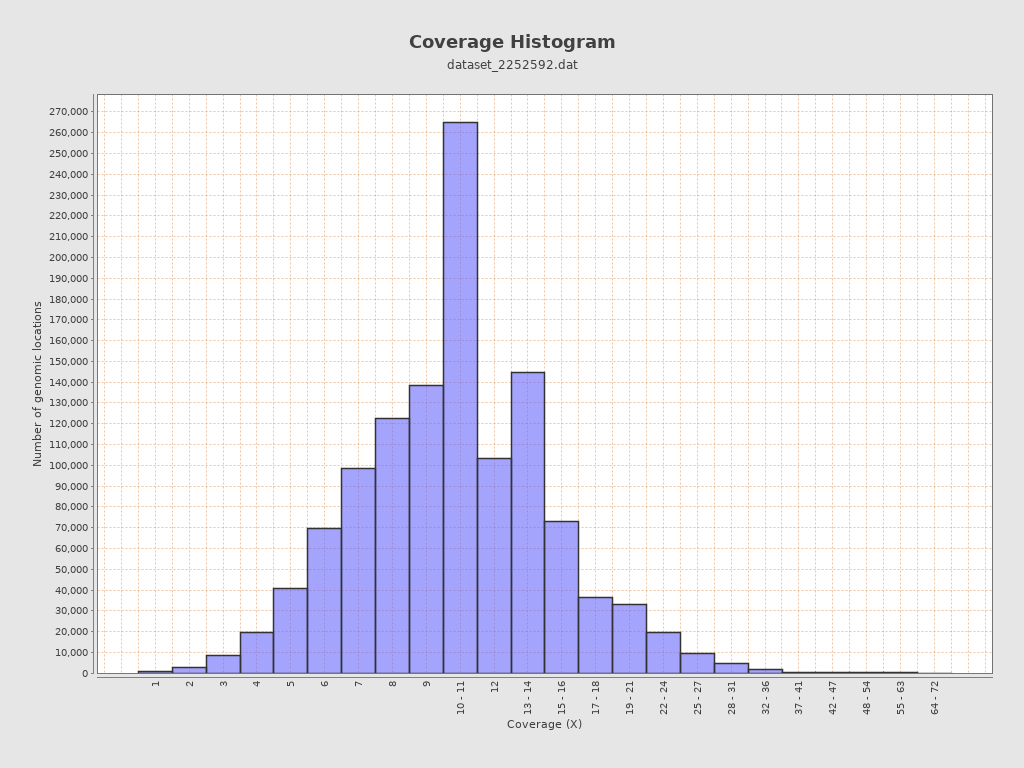
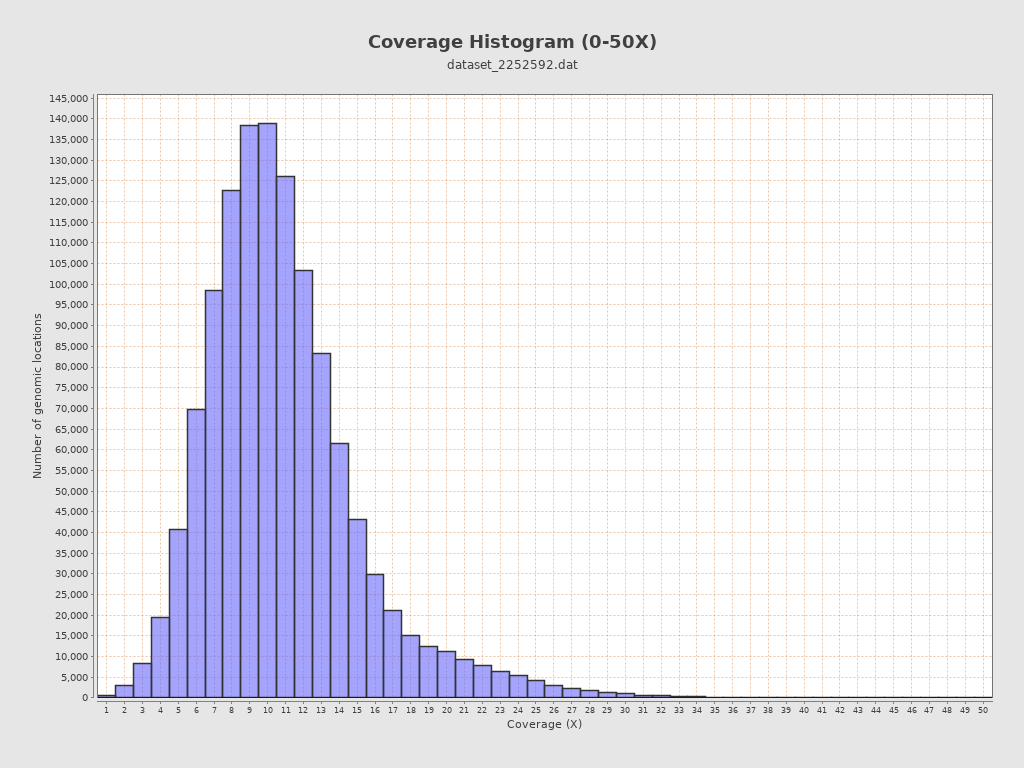
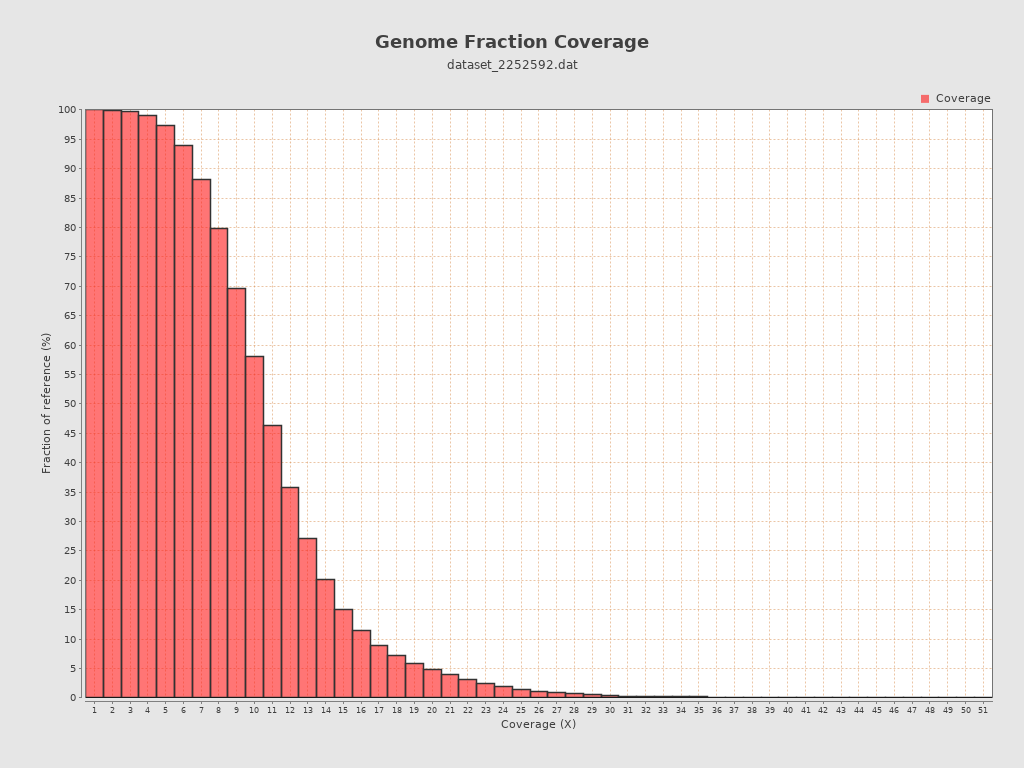
Coverage
| Mean | 10.8748 |
| Standard Deviation | 4.5201 |
| Mean (paired-end reads overlap ignored) | 10.85 |
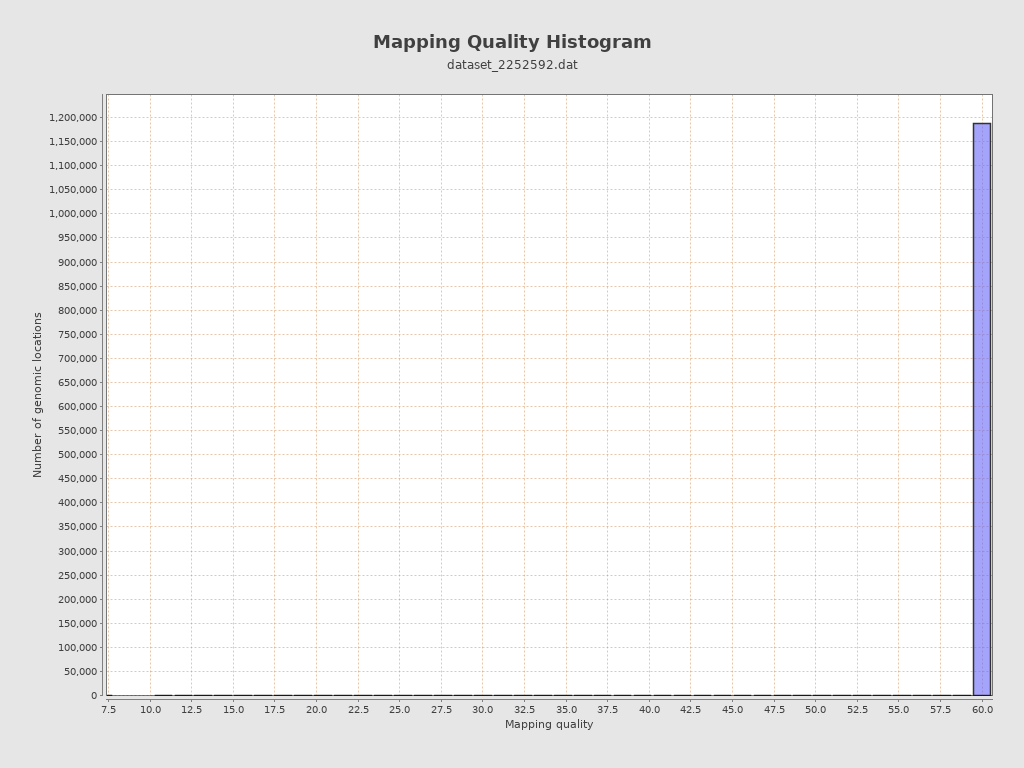
Mapping Quality
| Mean Mapping Quality | 59.6 |
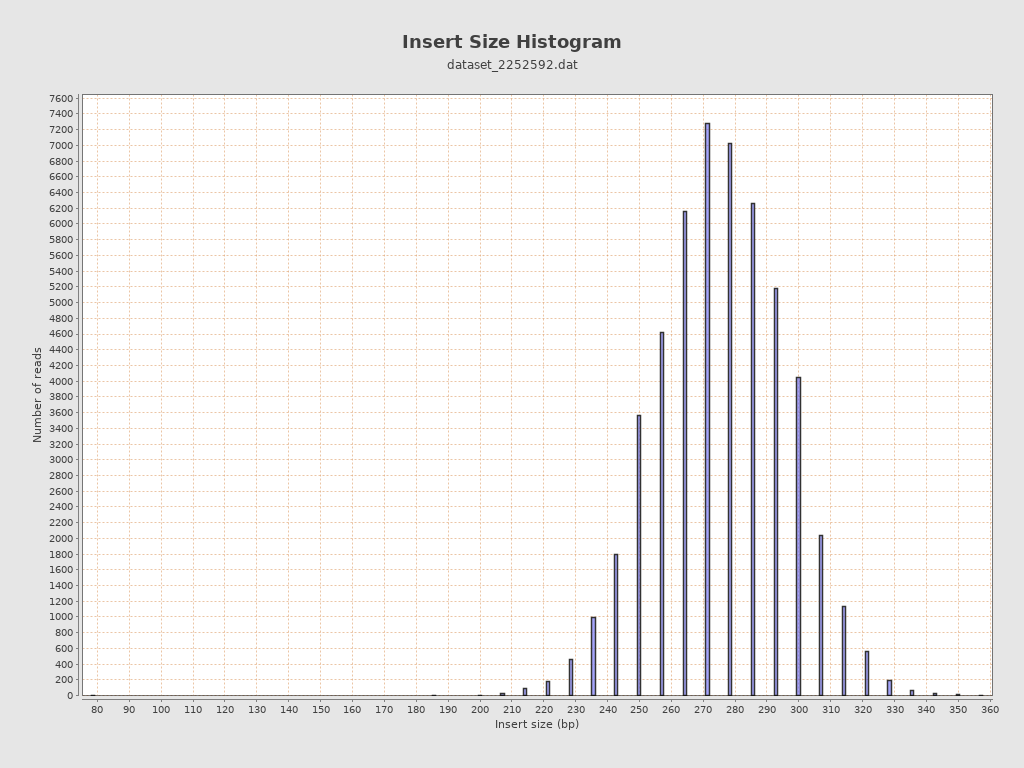
Insert size
| Mean | 279.27 |
| Standard Deviation | 19.95 |
| P25/Median/P75 | 266 / 279 / 293 |
Mismatches and indels
| General error rate | 0.18% |
| Mismatches | 23,767 |
| Insertions | 14 |
| Mapped reads with at least one insertion | 0.01% |
| Deletions | 20 |
| Mapped reads with at least one deletion | 0.02% |
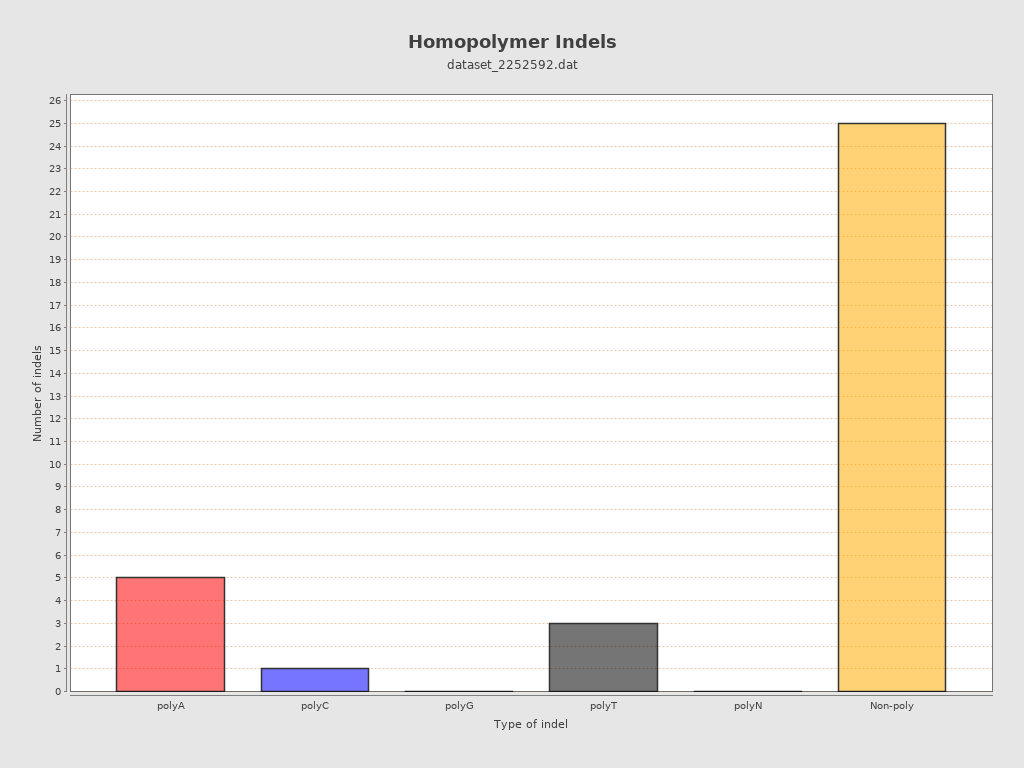
| Homopolymer indels | 26.47% |
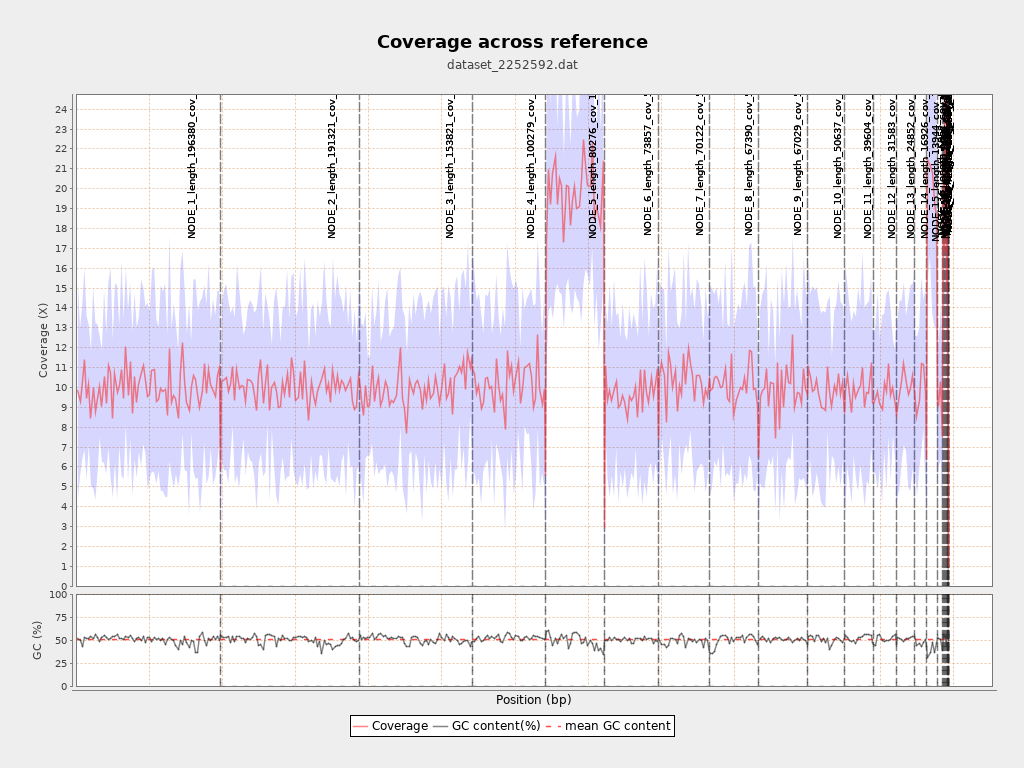
Chromosome stats
| Name | Length | Mapped bases | Mean coverage | Standard deviation |
| NODE_1_length_196380_cov_5.613936 | 196380 | 1967716 | 10.0199 | 3.1175 |
| NODE_2_length_191321_cov_5.603050 | 191321 | 1914764 | 10.0081 | 3.1004 |
| NODE_3_length_153821_cov_5.567414 | 153821 | 1529722 | 9.9448 | 3.2043 |
| NODE_4_length_100279_cov_5.685624 | 100279 | 1018117 | 10.1528 | 3.2375 |
| NODE_5_length_80276_cov_11.327483 | 80276 | 1592732 | 19.8407 | 4.5162 |
| NODE_6_length_73857_cov_5.433660 | 73857 | 716627 | 9.7029 | 3.0909 |
| NODE_7_length_70122_cov_5.782351 | 70122 | 722799 | 10.3077 | 3.0972 |
| NODE_8_length_67390_cov_5.684830 | 67390 | 683591 | 10.1438 | 3.1405 |
| NODE_9_length_67029_cov_5.560367 | 67029 | 665361 | 9.9265 | 3.2594 |
| NODE_10_length_50637_cov_5.532383 | 50637 | 499553 | 9.8654 | 3.0674 |
| NODE_11_length_39604_cov_5.671370 | 39604 | 393109 | 9.926 | 3.223 |
| NODE_12_length_31583_cov_5.433741 | 31583 | 305981 | 9.6882 | 2.8266 |
| NODE_13_length_24852_cov_5.646369 | 24852 | 250126 | 10.0646 | 3.1496 |
| NODE_14_length_16926_cov_5.635647 | 16926 | 167098 | 9.8723 | 3.1916 |
| NODE_15_length_13944_cov_11.422421 | 13944 | 283399 | 20.3241 | 4.2782 |
| NODE_16_length_6852_cov_5.406356 | 6852 | 65617 | 9.5763 | 3.0634 |
| NODE_17_length_1751_cov_6.142099 | 1751 | 18424 | 10.522 | 3.4083 |
| NODE_18_length_1466_cov_11.650602 | 1466 | 29080 | 19.8363 | 4.7663 |
| NODE_19_length_1332_cov_17.693031 | 1332 | 40250 | 30.2177 | 6.2551 |
| NODE_20_length_1213_cov_28.855786 | 1213 | 59326 | 48.9085 | 10.9403 |
| NODE_21_length_1124_cov_17.564079 | 1124 | 33572 | 29.8683 | 6.5459 |
| NODE_22_length_769_cov_11.106443 | 769 | 13785 | 17.9259 | 5.9356 |
| NODE_23_length_291_cov_10.322034 | 291 | 3900 | 13.4021 | 4.6089 |
| NODE_24_length_174_cov_6.714286 | 174 | 1002 | 5.7586 | 2.4281 |
| NODE_25_length_162_cov_5.504673 | 162 | 1249 | 7.7099 | 1.5543 |
| NODE_26_length_149_cov_5.436170 | 149 | 1094 | 7.3423 | 1.5273 |
| NODE_27_length_79_cov_20.333333 | 79 | 473 | 5.9873 | 0.1118 |
| NODE_28_length_63_cov_14.250000 | 63 | 63 | 1 | 0 |